2. Matched-filters¶
This notebook will provide a look at using EQcorrscan’s Tribe objects for matched-filter detection of earthquakes.
This notebook extends on the ideas covered in the Quick Start notebook. In particular this notebook also covers: 1. Concurrent execution of detection workflows for more efficient compute utilisation with large datasets; 2. Use of local waveform databases using obsplus; 3. Cross-correlation pick-correction using the lag_calc method.
2.1. Set up¶
We are going to focus in this notebook on using local data. For examples of how to directly use data from online providers see the Quick Start notebook.
To start off we will download some data - in your case this is likely data that you have either downloaded from one or more online providers, or data that you have collected yourself. At the moment we don’t care how those data are organised, as long as you have continuous data on disk somewhere. We will use obsplus to work out what data are were and provide us with the data that we need when we need it.
Obsplus is great and has more functionality than we expose here - if you make use of obsplus, please cite the paper by Chambers et al., (2021).
As in the Quick Start example, we will control the output level from EQcorrscan using logging.
[1]:
import logging
logging.basicConfig(
level=logging.WARNING,
format="%(asctime)s\t%(name)s\t%(levelname)s\t%(message)s")
Logger = logging.getLogger("TutorialLogger")
We will use the March 2023 Kawarau swarm as our case-study for this. This was an energetic swarm that was reported by New Zealand’s GeoNet monitoring agency and discussed in a news article here.
We will use data from ten stations over a duration of two days. The swarm lasted longer than this, but we need to limit compute resources for this tutorial! Feel free to change the end-date below to run for longer. To be kind to GeoNet and not repeatedly get data from their FDSN service we are going to get data from the AWS open data bucket. If you don’t already have boto3 installed you will need to install that for this sections (conda install boto3 or pip install boto3).
NB: If you actually want to access the GeoNet data bucket using Python, a drop-in replacement from FDSN clients exists here
[2]:
def get_geonet_data(starttime, endtime, stations, outdir):
import os
import boto3
from botocore import UNSIGNED
from botocore.config import Config
GEONET_AWS = "geonet-open-data"
DAY_STRUCT = "waveforms/miniseed/{date.year}/{date.year}.{date.julday:03d}"
CHAN_STRUCT = ("{station}.{network}/{date.year}.{date.julday:03d}."
"{station}.{location}-{channel}.{network}.D")
if not os.path.isdir(outdir):
os.makedirs(outdir)
bob = boto3.resource('s3', config=Config(signature_version=UNSIGNED))
s3 = bob.Bucket(GEONET_AWS)
date = starttime
while date < endtime:
day_path = DAY_STRUCT.format(date=date)
for station in stations:
for instrument in "HE":
for component in "ZNE12":
channel = f"{instrument}H{component}"
chan_path = CHAN_STRUCT.format(
station=station, network="NZ",
date=date, location="10", channel=channel)
local_path = os.path.join(outdir, chan_path)
if os.path.isfile(local_path):
Logger.info(f"Skipping {local_path}: exists")
continue
os.makedirs(os.path.dirname(local_path), exist_ok=True)
remote = "/".join([day_path, chan_path])
Logger.debug(f"Downloading from {remote}")
try:
s3.download_file(remote, local_path)
except Exception as e:
Logger.debug(f"Could not download {remote} due to {e}")
continue
Logger.info(f"Downloaded {remote}")
date += 86400
[3]:
%matplotlib inline
from obspy import UTCDateTime
starttime, endtime = UTCDateTime(2023, 3, 17), UTCDateTime(2023, 3, 19)
stations = ['EDRZ', 'LIRZ', 'MARZ', 'MKRZ', 'OMRZ', 'OPRZ', 'TARZ', 'WKHS', 'HNCZ', 'KARZ']
outdir = "tutorial_waveforms"
get_geonet_data(starttime=starttime, endtime=endtime, stations=stations, outdir=outdir)
Great, now we have some data. EQcorrscan is well set up to use clients for data access, using clients allows EQcorrscan to request the data that it needs and take care of overlapping chunks of data to ensure that no data are missed: network-based matched-filters apply a delay-and-stack step to the correlations from individual channels. This increases the signal-to-noise ratio of the correlation sum. However, because of the delay part, the stacks made at start and end of chunks of waveform data do not use the full network. To get around this you should overlap your data.
If you use client-based access to data, EQcorrscan will take care of this for you.
So how do you use clients for local data? Make a local database using obsplus.
If you don’t have obsplus installed you should install it now (conda install obsplus or pip install obsplus).
[4]:
from obsplus import WaveBank
bank = WaveBank(outdir)
bank.get_availability_df()
[4]:
| network | station | location | channel | starttime | endtime | |
|---|---|---|---|---|---|---|
| 0 | NZ | EDRZ | 10 | EHE | 2023-03-17 00:00:03.528394 | 2023-03-19 00:00:00.098393 |
| 1 | NZ | EDRZ | 10 | EHN | 2023-03-17 00:00:05.458394 | 2023-03-19 00:00:04.518393 |
| 2 | NZ | EDRZ | 10 | EHZ | 2023-03-17 00:00:03.528394 | 2023-03-19 00:00:03.588393 |
| 3 | NZ | KARZ | 10 | EHE | 2023-03-17 00:00:02.963130 | 2023-03-19 00:00:01.273126 |
| 4 | NZ | KARZ | 10 | EHN | 2023-03-17 00:00:00.093130 | 2023-03-19 00:00:00.303126 |
| 5 | NZ | KARZ | 10 | EHZ | 2023-03-17 00:00:05.823130 | 2023-03-19 00:00:03.653126 |
| 6 | NZ | LIRZ | 10 | EHE | 2023-03-17 00:00:01.753132 | 2023-03-19 00:00:03.523130 |
| 7 | NZ | LIRZ | 10 | EHN | 2023-03-17 00:00:02.913132 | 2023-03-19 00:00:04.253130 |
| 8 | NZ | LIRZ | 10 | EHZ | 2023-03-17 00:00:01.463132 | 2023-03-19 00:00:00.313130 |
| 9 | NZ | MARZ | 10 | EHE | 2023-03-17 00:00:01.553130 | 2023-03-19 00:00:01.593131 |
| 10 | NZ | MARZ | 10 | EHN | 2023-03-17 00:00:01.683130 | 2023-03-19 00:00:04.163131 |
| 11 | NZ | MARZ | 10 | EHZ | 2023-03-17 00:00:00.963130 | 2023-03-19 00:00:05.063131 |
| 12 | NZ | MKRZ | 10 | EHE | 2023-03-17 00:00:01.673129 | 2023-03-19 00:00:01.763133 |
| 13 | NZ | MKRZ | 10 | EHN | 2023-03-17 00:00:00.143129 | 2023-03-19 00:00:02.463133 |
| 14 | NZ | MKRZ | 10 | EHZ | 2023-03-17 00:00:00.053129 | 2023-03-19 00:00:02.363133 |
| 15 | NZ | OMRZ | 10 | EHE | 2023-03-17 00:00:02.740000 | 2023-03-19 00:00:02.470000 |
| 16 | NZ | OMRZ | 10 | EHN | 2023-03-17 00:00:00.580000 | 2023-03-19 00:00:00.550000 |
| 17 | NZ | OMRZ | 10 | EHZ | 2023-03-17 00:00:04.110000 | 2023-03-19 00:00:03.820000 |
| 18 | NZ | OPRZ | 10 | HHE | 2023-03-17 00:00:02.993132 | 2023-03-19 00:00:01.243131 |
| 19 | NZ | OPRZ | 10 | HHN | 2023-03-17 00:00:03.473132 | 2023-03-19 00:00:04.443131 |
| 20 | NZ | OPRZ | 10 | HHZ | 2023-03-17 00:00:01.963132 | 2023-03-19 00:00:00.143131 |
| 21 | NZ | TARZ | 10 | EHE | 2023-03-17 00:00:01.850000 | 2023-03-19 00:00:01.580000 |
| 22 | NZ | TARZ | 10 | EHN | 2023-03-17 00:00:00.760000 | 2023-03-19 00:00:00.820000 |
| 23 | NZ | TARZ | 10 | EHZ | 2023-03-17 00:00:00.630000 | 2023-03-19 00:00:03.830000 |
Obsplus has now scanned the waveforms that we just downloaded and made a table of what is there. Great. These WaveBank objects have a similar api to obspy Client objects, so we can use them as a drop-in replacement.
Now we are nearly ready to make some templates.
2.2. Template creation¶
To make templates you need two things: 1. Continuous waveform data; 2. A catalogue of events with picks.
We already have (1). For (2), the catalogue of events, we could use GeoNet picked events, however if you have events with picks locally and want to use those events as templates you should save those events in a format readable by obspy. You can then skip ahead to read those picks back in.
In the worst case scenario you have times that you know that you want your template to start at, but they are not in any standard format readable by obspy, you can construct events from scratch as below. Note in this example I am just populating the picks as this is all we need. You do need to be careful about the waveform_id: this should match the seed id of the continuous data exactly, otherwise the picks will not be used.
[5]:
from obspy.core.event import (
Catalog, Event, Pick, WaveformStreamID)
from obspy import UTCDateTime
# Make the picks for the event:
picks = [
Pick(
time=UTCDateTime(2023, 3, 18, 7, 46, 15, 593125),
waveform_id=WaveformStreamID(
network_code='NZ', station_code='MARZ',
channel_code='EHZ', location_code='10'),
phase_hint='P'),
Pick(
time=UTCDateTime(2023, 3, 18, 7, 46, 17, 633115),
waveform_id=WaveformStreamID(
network_code='NZ', station_code='MKRZ',
channel_code='EHZ', location_code='10'),
phase_hint='P'),
Pick(
time=UTCDateTime(2023, 3, 18, 7, 46, 18, 110000),
waveform_id=WaveformStreamID(
network_code='NZ', station_code='OMRZ',
channel_code='EHZ', location_code='10'),
phase_hint='P'),
]
# Add as many picks as you have - you might want to loop
# and/or make a function to pasre your picks to obspy Picks.
# Make the event
event = Event(picks=picks)
# Make the catalog
catalog = Catalog([event])
For this example we are going to use a catalogue of events picked by GeoNet - we will download those data and write them to disk to mimic you using local files:
[6]:
from obspy.clients.fdsn import Client
client = Client("GEONET")
cat = client.get_events(
starttime=UTCDateTime(2023, 3, 17),
endtime=UTCDateTime(2023, 3, 19),
latitude=-38.05, longitude=176.73,
maxradius=0.5, minmagnitude=3.0) # Limited set of relevent events
cat.write("tutorial_catalog.xml", format="QUAKEML")
2.3. Template creation with local files¶
Now that we have the events and waveforms we need, we can make our Tribe of templates.
First we have to read in the events that we want to use as templates:
[7]:
from obspy import read_events
cat = read_events("tutorial_catalog.xml")
print(cat)
49 Event(s) in Catalog:
2023-03-17T14:29:34.921582Z | -38.067, +176.689 | 3.40 MLv | manual
2023-03-17T14:56:17.215087Z | -38.061, +176.679 | 3.07 MLv | manual
...
2023-03-18T20:20:52.842474Z | -38.045, +176.734 | 3.17 MLv | manual
2023-03-18T21:42:39.943071Z | -38.051, +176.735 | 4.25 MLv | manual
To see all events call 'print(CatalogObject.__str__(print_all=True))'
2.3.1. Pick curation¶
You may want to limit what picks you actually use for your templates. Any picks that you provide will be used for cutting waveforms - this may include amplitude picks! You should not need to restrict what stations you have picks for, but it doesn’t do any harm to.
Below we select picks from the stations that we set earlier, and only P and S picks. We also limit to only one P and one S pick per station - you may not want to do that, but it can get messy if you have multiple picks of the same phase.
[8]:
from eqcorrscan.utils.catalog_utils import filter_picks
cat = filter_picks(
cat,
stations=stations,
phase_hints=["P", "S"],
enforce_single_pick="earliest")
Matched-filter GPU is not compiled! Should be here: /home/chambeca/my_programs/Building/fast_matched_filter/fast_matched_filter/lib/matched_filter_GPU.so
2.3.2. Template creation decisions¶
We now have everything needed to create a tribe of templates. At this point you have to make some decisions about parameters: 1. What filters to use; 2. What sampling-rate to use; 3. How long your template waveforms should be; 4. When to start your template waveforms relative to your pick times; 5. Whether to use separate P and S windows or not.
Your choices for 2 and 3 should be based somewhat on your choice of what filters to use (1). There is little point in using a sampling-rate significantly above 2.5x your high-cut frequency (2.5x because off the roll-offs used in the Butterworth filters used by EQcorrscan). Lower sampling-rates will result in fewer correlations and hence faster compute time, but most of the time in EQcorrscan’s matched-filter runs is spent in the pre-processing of the data rather than the actual correlation computation.
When deciding on filtering parameters you may find it helpful to look at what frequencies have the best signal-to-noise ratio. There are functions in the eqcorrscan.utils.plotting module to help with this.
We will use some relatively standard, but un-tuned parameters for this example. You should spend some time deciding: these decisions strongly affect the quality of your results. You can also set the minimum signal-to-noise ratio for traces to be included in your templates. Again, this should be tested.
The process_len parameter controls how much data will be processed at once. Because EQocrrscan computes resampling in the frequency domain, and can compute correlations in the frequency domain, changing this length between construction and detection affects the accuracy of the Fourier transforms which affects the final correlations. For this reason the process_len is maintained throughout the workflow by EQcorrscan. Here we use one hour (3600 seconds), but it is common to use one day
(86400 seconds).
You will note that we use the from_client method for construction: this is because we have a WaveBank that emulates a client making this really simple.
[9]:
from eqcorrscan import Tribe
tribe = Tribe().construct(
method="from_client",
client_id=bank,
catalog=cat,
lowcut=2.0,
highcut=15.0,
samp_rate=50.0,
filt_order=4,
length=3.0,
prepick=0.5,
swin="all",
process_len=3600,
all_horix=True,
min_snr=4.0,
parallel=True
)
2023-12-01 13:20:51,205 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 2.876741406237065 below threshold for KARZ.EHZ, not using
2023-12-01 13:20:51,206 eqcorrscan.core.template_gen WARNING No pick for NZ.KARZ.10.EHZ
2023-12-01 13:20:51,208 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 1.9540223699044974 below threshold for OMRZ.EHZ, not using
2023-12-01 13:20:51,209 eqcorrscan.core.template_gen WARNING No pick for NZ.OMRZ.10.EHZ
2023-12-01 13:20:51,274 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 1.5737322242199878 below threshold for LIRZ.EHZ, not using
2023-12-01 13:20:51,275 eqcorrscan.core.template_gen WARNING No pick for NZ.LIRZ.10.EHZ
2023-12-01 13:20:51,277 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 0.8545790599562736 below threshold for MKRZ.EHN, not using
2023-12-01 13:20:51,278 eqcorrscan.core.template_gen WARNING No pick for NZ.MKRZ.10.EHN
2023-12-01 13:20:51,280 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 1.5224417059935123 below threshold for OMRZ.EHZ, not using
2023-12-01 13:20:51,280 eqcorrscan.core.template_gen WARNING No pick for NZ.OMRZ.10.EHZ
2023-12-01 13:20:51,283 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 0.9704676413162124 below threshold for TARZ.EHZ, not using
2023-12-01 13:20:51,283 eqcorrscan.core.template_gen WARNING No pick for NZ.TARZ.10.EHZ
2023-12-01 13:20:51,315 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 3.6693293207326403 below threshold for LIRZ.EHZ, not using
2023-12-01 13:20:51,316 eqcorrscan.core.template_gen WARNING No pick for NZ.LIRZ.10.EHZ
2023-12-01 13:20:51,318 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 3.241518566731996 below threshold for OMRZ.EHZ, not using
2023-12-01 13:20:51,319 eqcorrscan.core.template_gen WARNING No pick for NZ.OMRZ.10.EHZ
2023-12-01 13:20:51,322 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 2.657470431167782 below threshold for TARZ.EHZ, not using
2023-12-01 13:20:51,322 eqcorrscan.core.template_gen WARNING No pick for NZ.TARZ.10.EHZ
2023-12-01 13:20:52,446 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 0.5290305215816998 below threshold for LIRZ.EHZ, not using
2023-12-01 13:20:52,446 eqcorrscan.core.template_gen WARNING No pick for NZ.LIRZ.10.EHZ
2023-12-01 13:20:52,448 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 1.0144665058782114 below threshold for TARZ.EHZ, not using
2023-12-01 13:20:52,448 eqcorrscan.core.template_gen WARNING No pick for NZ.TARZ.10.EHZ
2023-12-01 13:20:53,107 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 2.400855581315436 below threshold for LIRZ.EHZ, not using
2023-12-01 13:20:53,108 eqcorrscan.core.template_gen WARNING No pick for NZ.LIRZ.10.EHZ
2023-12-01 13:20:53,109 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 2.3176851693587057 below threshold for MKRZ.EHZ, not using
2023-12-01 13:20:53,110 eqcorrscan.core.template_gen WARNING No pick for NZ.MKRZ.10.EHZ
2023-12-01 13:20:53,112 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 1.3894705992621912 below threshold for OMRZ.EHE, not using
2023-12-01 13:20:53,112 eqcorrscan.core.template_gen WARNING No pick for NZ.OMRZ.10.EHE
2023-12-01 13:20:53,114 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 3.884291218472121 below threshold for OPRZ.HHZ, not using
2023-12-01 13:20:53,115 eqcorrscan.core.template_gen WARNING No pick for NZ.OPRZ.10.HHZ
2023-12-01 13:20:53,173 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 3.1825458574844174 below threshold for MARZ.EHN, not using
2023-12-01 13:20:53,174 eqcorrscan.core.template_gen WARNING No pick for NZ.MARZ.10.EHN
2023-12-01 13:20:53,177 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 3.851222353660108 below threshold for OPRZ.HHN, not using
2023-12-01 13:20:53,178 eqcorrscan.core.template_gen WARNING No pick for NZ.OPRZ.10.HHN
2023-12-01 13:20:53,697 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 0.4135717198623312 below threshold for KARZ.EHZ, not using
2023-12-01 13:20:53,697 eqcorrscan.core.template_gen WARNING No pick for NZ.KARZ.10.EHZ
2023-12-01 13:20:53,699 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 2.3877145548728342 below threshold for LIRZ.EHE, not using
2023-12-01 13:20:53,699 eqcorrscan.core.template_gen WARNING No pick for NZ.LIRZ.10.EHE
2023-12-01 13:20:53,701 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 1.1370695659576338 below threshold for MARZ.EHZ, not using
2023-12-01 13:20:53,701 eqcorrscan.core.template_gen WARNING No pick for NZ.MARZ.10.EHZ
2023-12-01 13:20:53,703 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 0.22436151966111806 below threshold for OMRZ.EHZ, not using
2023-12-01 13:20:53,704 eqcorrscan.core.template_gen WARNING No pick for NZ.OMRZ.10.EHZ
2023-12-01 13:20:53,705 eqcorrscan.core.template_gen WARNING Signal-to-noise ratio 1.487347592301636 below threshold for OPRZ.HHZ, not using
2023-12-01 13:20:53,706 eqcorrscan.core.template_gen WARNING No pick for NZ.OPRZ.10.HHZ
2023-12-01 13:20:57,350 eqcorrscan.core.match_filter.tribe ERROR Empty Template
2023-12-01 13:20:57,352 eqcorrscan.core.match_filter.tribe ERROR Empty Template
You should see an ERROR message about empty templates: some of the events in our catalog do not have useful data in our wavebank. We might want to set a minimum number of stations used for our templates to ensure that our templates are all of reasonable quality. In this case we will only retain templates with at least five stations:
[10]:
tribe.templates = [t for t in tribe if len({tr.stats.station for tr in t.st}) >= 5]
print(tribe)
Tribe of 33 templates
2.3.3. Matched-filter detection¶
Now that we have our tribe we can use it to detect new earthquakes. Again we will make use of our local WaveBank. This is preferred to feeding one stream of data to the code at a time for two reasons: 1. EQcorrscan will overlap your chunks of data (in this can every hour of data) to ensure that all of the data have correlations from all channels after the delay-and-stack correlation sums. 2. EQcorrscan can pre-emptively process the next chunks data in parallel while computing detections in
the current chunk. This can significantly speed up processing, and makes better use of compute resources.
The main decisions that you have to make at this stage are around thresholds. Generally it is better to start with a relatively low threshold: you can increase the threshold later using the Party.rethreshold method, but you can’t lower it without re-running the whole detection workflow.
It is common to use MAD thresholding, but you should experiment with your dataset to see what works best.
[11]:
party = tribe.client_detect(
client=bank,
starttime=starttime,
endtime=endtime,
threshold=10.0,
threshold_type="MAD",
trig_int=1.0,
)
2023-12-01 13:21:02,031 eqcorrscan.core.match_filter.tribe WARNING Using concurrent_processing=True can be faster ifdownloading your data takes a long time. See https://github.com/eqcorrscan/EQcorrscan/pull/544for benchmarks.
2023-12-01 13:22:27,562 eqcorrscan.core.match_filter.helpers.tribe WARNING Removed data for NZ.EDRZ.10.EHE NZ.EDRZ.10.EHN NZ.EDRZ.10.EHZ NZ.KARZ.10.EHE NZ.KARZ.10.EHN NZ.KARZ.10.EHZ NZ.LIRZ.10.EHE NZ.LIRZ.10.EHN NZ.LIRZ.10.EHZ NZ.MARZ.10.EHE NZ.MARZ.10.EHN NZ.MARZ.10.EHZ NZ.MKRZ.10.EHE NZ.MKRZ.10.EHN NZ.MKRZ.10.EHZ NZ.OMRZ.10.EHE NZ.OMRZ.10.EHN NZ.OMRZ.10.EHZ NZ.OPRZ.10.HHE NZ.OPRZ.10.HHN NZ.OPRZ.10.HHZ NZ.TARZ.10.EHN NZ.TARZ.10.EHZ due to less than 80% of the required length.
2023-12-01 13:22:27,563 eqcorrscan.core.match_filter.tribe WARNING No suitable data between 2023-03-18T23:50:43.199856Z and 2023-03-19T00:51:03.199856Z, skipping
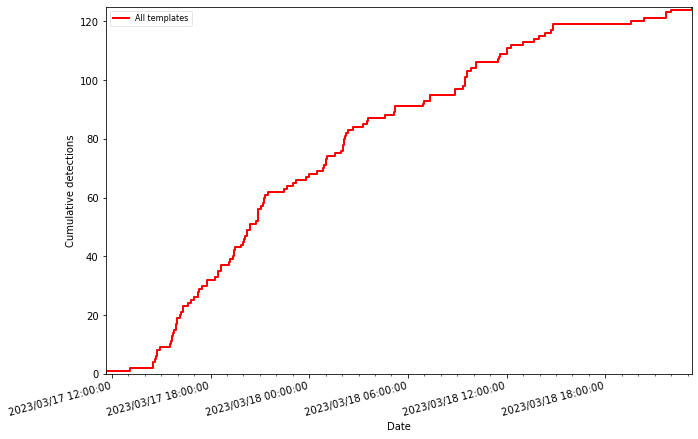
[12]:
print(len(party))
125
[13]:
party.plot(plot_grouped=True)
2.4. Note on concurrent processing¶
As of EQcorrscan versions > 0.4.4, detect methods support concurrent processing of intermediate steps in the matched-filter process when running multiple chunks of data (e.g. when endtime - starttime > process_len). By default this is disabled as it does increase memory use. However, for cases when downloading data from a client is a major bottleneck, or processing data is slow (e.g. when you have limited CPU threads available, but do have a GPU for the FMF correlation backend), and can cope
with the extra memory requirements, this can be much faster.
To see examples of the speed-ups and memory consumption, look at the benchmarks in the pull-request here.
To enable concurrent processing, use the concurrent_processing argument for .client_detect or .detect methods on Tribe objects.
[ ]: